PLASMODESMA, NMR-based deconvolution method
CASC4DE has contributed to the development of Plasmodesma, an automated NMR-based pharmacophoric deconvolution method for plant extracts. It has also set up a dedicated data analysis server.
This technique allows to reduce considerably the time needed to identify active compounds from natural extracts. Thus it represents a non-negligible resource for drug discovery.
This method is based on the automatic acquisition of a standard set of NMR experiments from a medium-sized set of samples differing by their bioactivity. From this, a pipeline is run and the data is analysed by leveraging machine learning approaches in order to extract the molecular spectral fingerprints of the active compounds from unrefined fractions of a crude extract.
About Plasmodesma method and web service:
-
Regarding the analytical method itself : L. Margueritte, P. Markov, L. Chiron, J.-P. Starck, C. Vonthron-Sénécheau, M. Bourjot & M.-A. Delsuc, « Automatic differential analysis of NMR experiments in complex samples. » Magn. Reson. Chem., (2018), 56, 469–479.
-
A simplified Plasmodesma data analysis service is available freely on the internet http://plasmodesma.igbmc.science (under the Cecill A licence) and has an interactive user-friendly graphic interface. Read more on Faraday Discuss., 2019,218, 441-458 10.1039/C8FD00242H
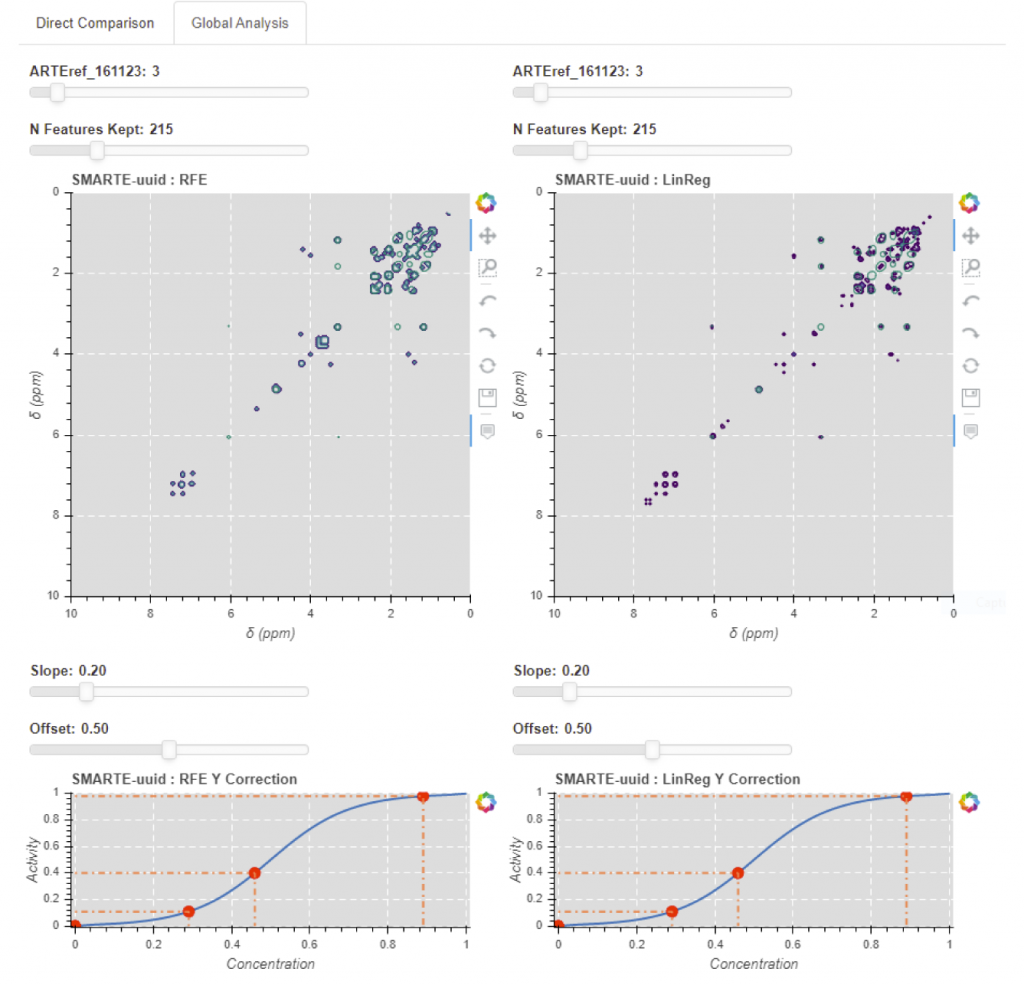
Interactive display of a correlation analysis obtained from a whole set of data from the measured activity response. 1. The top view presents the results of the Recursive Feature Elimination method (left) and of the linear regression (right). The number of selected features can be varied, thus varying the number of displayed spectral channels. 2. Both bottom view present the activity to concentration estimator function, with controls for the slope and offset.
Plasmodesma code is available at: https://github.com/delsuc/plasmodesma




