Versatile decision-making tool for NMR spectral assignments of proteins
CASC4DE has developped RESCUE 3 in collaboration with Thérèse Malliavin from Université de Lorraine and Marc-André Delsuc from IGBMC.
Spectral assignments of proteins in NMR is crucial as, even today, this generally requires lots of manual work and good expertise. The development of automatic techniques would therefore greatly accelerate analyses and thus enables a higher throughput in cases where this step is required, such as for ligand interaction screening, protein-protein recognition, NMR structure determination and/or when assignment information is not obtainable (e.g non labeled proteins, methyl-based approaches).
RESCUE is a statistical approach developed in 1999 for NMR spectral assignment of proteins through a simple artificial neural network (perceptron) [1]. It allowed the type of amino acid to be determined from the observed 1H chemical shift. It used as a training set the data available in the BMRB (bmrb.io) at the time. It was extended in 2004 to the whole spin set, using a “Naive Bayes” probabilistic model for each amino acid type from a set of given chemical shifts [2]. Predictions accuracy of this second version reaches up to 75%.
The recent development in Deep Learning (DL) as well as the current size of the BMRB database available today allow to get better assignment predictions. We developed a Deep Neural Network (DNN) together with a cleaned database by removing duplicates, strongly homolog entries, non-protein sequences and scarcely assign entries and by realigning chemical shift references. We obtain a final database from the BMRB including of 485264 sets of chemical shifts and a representation of the 20 classical amino acids to feed the neural network.
We developed a 7 dense layers neural network using the open-source Keras Python library. An adamax optimizer and a categorical crossentropy loss were used, with training on 25 epochs. We then built scenarios to filter the chemical shift sets according to what is acquired experimentally. This allows us to adapt the DNN to each sets of available experiments. The newly developed algorithm has been tested in several situations reproducing possible scenarios and allowing to evaluate the efficiency of the program in different cases. In all cases the algorithm shows very good results and is able to make relevant amino acid predictions with assessment of prediction accuracy.
This work will soon be published and available to deployment adapted to labs or companies needs.
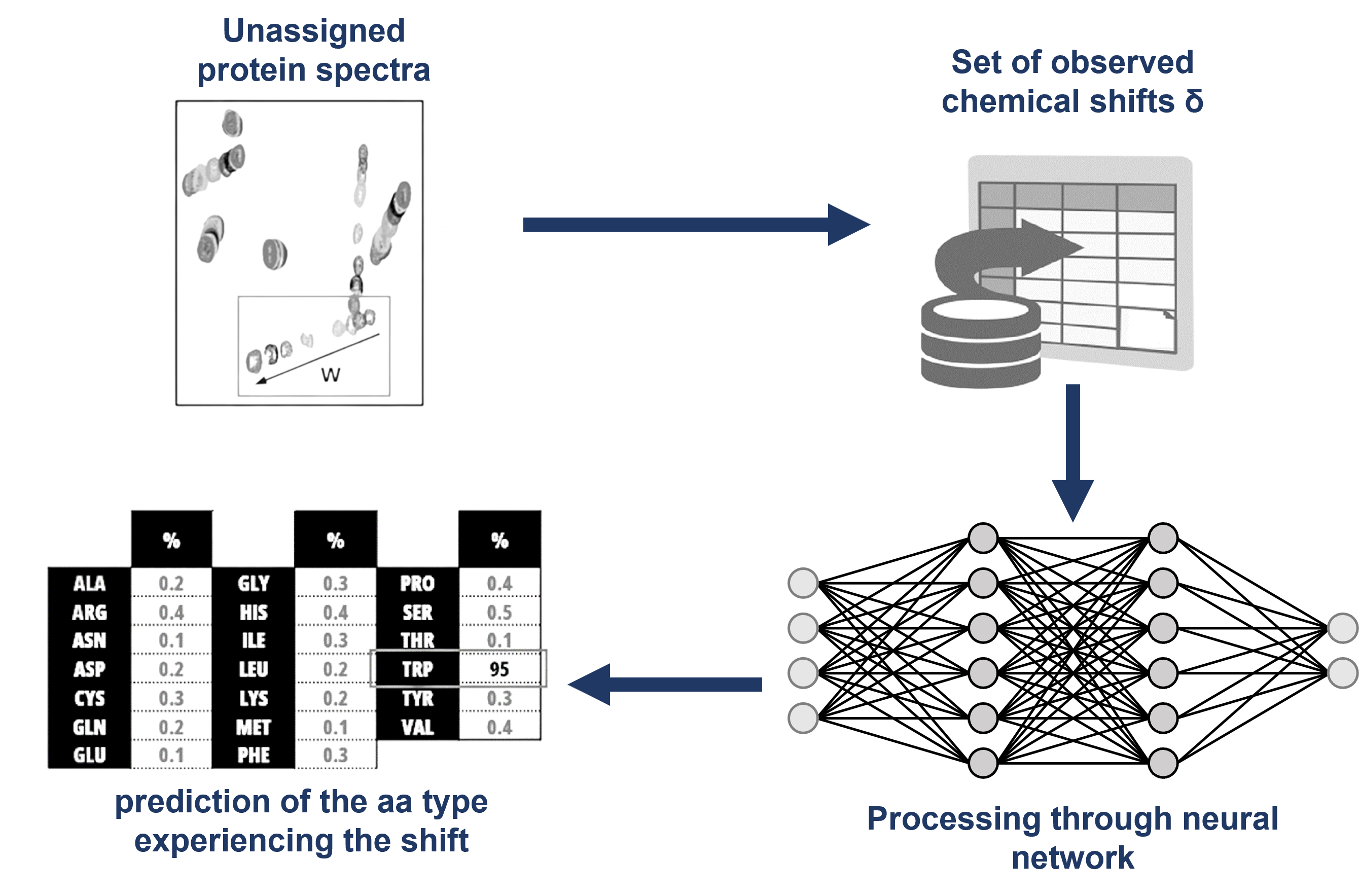
The RESCUE 3 approach.
Refs.:
- [1] J. L. Pons and M. A. Delsuc J. Biomol. NMR 15 (1), 15 26 (1999)
- [2] A. Marin, T. E. Malliavin, P. Nicolas, and M.-A. Delsuc. J. Biomol. NMR 30 (1), 47 60 (2004)




